Clustering Comparison: Leiden vs Louvain
[1]:
import os
import bokeh
from bokeh.plotting import show
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import phenograph
import pacmap
from sklearn.preprocessing import StandardScaler, MinMaxScaler
import flowkit as fk
bokeh.io.output_notebook()
%matplotlib inline
Load Samples & FlowJo 10 workspace
[2]:
base_dir = "../../../data/8_color_data_set/"
sample_path = os.path.join(base_dir, "fcs_files")
wsp_path = os.path.join(base_dir, "8_color_ICS.wsp")
seed = 123
[3]:
workspace = fk.Workspace(wsp_path, fcs_samples=sample_path)
[4]:
sample_groups = workspace.get_sample_groups()
sample_groups
[4]:
['All Samples', 'DEN', 'GEN', 'G69', 'Lyo Cells']
[5]:
sample_group = 'DEN'
[6]:
sample_ids = workspace.get_sample_ids()
sample_ids
[6]:
['101_DEN084Y5_15_E01_008_clean.fcs',
'101_DEN084Y5_15_E03_009_clean.fcs',
'101_DEN084Y5_15_E05_010_clean.fcs']
[7]:
sample_id = '101_DEN084Y5_15_E01_008_clean.fcs'
[8]:
print(workspace.get_gate_hierarchy(sample_id, output='ascii'))
root
╰── Time
╰── Singlets
╰── aAmine-
╰── CD3+
├── CD4+
│ ├── CD107a+
│ ├── IFNg+
│ ├── IL2+
│ ╰── TNFa+
╰── CD8+
├── CD107a+
├── IFNg+
├── IL2+
╰── TNFa+
Run analyze_samples & retrieve gated events as DataFrames
[9]:
workspace.analyze_samples(sample_group)
[10]:
dfs = []
for sample_id in sample_ids:
df = workspace.get_gate_events(sample_id, gate_name="Singlets")
dfs.append(df)
[11]:
dfs[0].head()
[11]:
| sample_id | FSC-A | FSC-H | FSC-W | SSC-A | SSC-H | SSC-W | TNFa FITC FLR-A | CD8 PerCP-Cy55 FLR-A | IL2 BV421 FLR-A | Aqua Amine FLR-A | IFNg APC FLR-A | CD3 APC-H7 FLR-A | CD107a PE FLR-A | CD4 PE-Cy7 FLR-A | Time | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.632765 | 0.519402 | 0.304564 | 0.116014 | 0.111382 | 0.260397 | 0.253992 | 0.225618 | 0.253962 | 0.250438 | 0.235338 | 0.419341 | 0.276203 | 0.548099 | 0.036353 |
| 7 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.428379 | 0.338997 | 0.315916 | 0.205931 | 0.192833 | 0.266981 | 0.241998 | 0.240605 | 0.328635 | 0.248555 | 0.241893 | 0.240895 | 0.283410 | 0.256122 | 0.036381 |
| 8 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.600745 | 0.502071 | 0.299133 | 0.349897 | 0.309628 | 0.282515 | 0.556043 | 0.324994 | 0.397557 | 0.305441 | 0.272172 | 0.244708 | 0.701451 | 0.301256 | 0.036396 |
| 9 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.415333 | 0.329010 | 0.315593 | 0.200316 | 0.182648 | 0.274184 | 0.254298 | 0.329108 | 0.320049 | 0.257477 | 0.271226 | 0.500196 | 0.319537 | 0.594672 | 0.036452 |
| 10 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.427080 | 0.328156 | 0.325364 | 0.296132 | 0.262680 | 0.281837 | 0.260209 | 0.296330 | 0.316296 | 0.262380 | 0.266253 | 0.451234 | 0.284111 | 0.618065 | 0.036467 |
[12]:
# subsample 10k events and ignore sample_id & Time columns
k = 10_000
X = pd.concat([df.iloc[:, 1:-1].sample(k) for df in dfs])
[13]:
X.head()
[13]:
| FSC-A | FSC-H | FSC-W | SSC-A | SSC-H | SSC-W | TNFa FITC FLR-A | CD8 PerCP-Cy55 FLR-A | IL2 BV421 FLR-A | Aqua Amine FLR-A | IFNg APC FLR-A | CD3 APC-H7 FLR-A | CD107a PE FLR-A | CD4 PE-Cy7 FLR-A | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25932 | 0.225668 | 0.176079 | 0.320408 | 0.151519 | 0.140053 | 0.270468 | 0.247182 | 0.308449 | 0.295386 | 0.238875 | 0.240529 | 0.530728 | 0.267461 | 0.579704 |
| 363 | 0.460844 | 0.404091 | 0.285112 | 0.081598 | 0.080498 | 0.253417 | 0.260949 | 0.295048 | 0.455798 | 0.560543 | 0.259499 | 0.470362 | 0.304936 | 0.532702 |
| 148278 | 0.471904 | 0.396152 | 0.297805 | 0.305392 | 0.288380 | 0.264748 | 0.274476 | 0.266639 | 0.444418 | 0.518590 | 0.255009 | 0.220964 | 0.537843 | 0.310567 |
| 26687 | 0.549719 | 0.444595 | 0.309112 | 0.192974 | 0.185135 | 0.260585 | 0.241193 | 0.310688 | 0.282620 | 0.264474 | 0.262676 | 0.471560 | 0.241348 | 0.577387 |
| 100730 | 0.655915 | 0.530857 | 0.308895 | 0.125434 | 0.118504 | 0.264620 | 0.252927 | 0.574966 | 0.276928 | 0.249968 | 0.248684 | 0.422043 | 0.266377 | 0.204461 |
Perform Louvain & Leiden clustering
[14]:
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
[15]:
communities_louvain, graph_louvain, Q_louvain = phenograph.cluster(
X_scaled,
clustering_algo='louvain',
seed=seed
)
Finding 30 nearest neighbors using minkowski metric and 'auto' algorithm
Neighbors computed in 4.576421737670898 seconds
Jaccard graph constructed in 0.7761034965515137 seconds
Wrote graph to binary file in 0.17565011978149414 seconds
Running Louvain modularity optimization
After 1 runs, maximum modularity is Q = 0.853431
After 3 runs, maximum modularity is Q = 0.856286
Louvain completed 23 runs in 11.92734169960022 seconds
Sorting communities by size, please wait ...
PhenoGraph completed in 17.65075969696045 seconds
[16]:
communities_leiden, graph_leiden, Q_leiden = phenograph.cluster(
X_scaled,
clustering_algo='leiden',
seed=seed
)
Finding 30 nearest neighbors using minkowski metric and 'auto' algorithm
Neighbors computed in 3.6213481426239014 seconds
Jaccard graph constructed in 0.942486047744751 seconds
Running Leiden optimization
Leiden completed in 8.978180170059204 seconds
Sorting communities by size, please wait ...
PhenoGraph completed in 14.09741997718811 seconds
[17]:
titles = ['Leiden', 'Louvain']
communities = [communities_leiden, communities_louvain]
[18]:
leiden_means = [
X_scaled[communities_leiden==i, :].mean(axis=0)
for i in np.unique(communities_leiden)
]
leiden_clusters = pd.DataFrame(
leiden_means,
columns = X.columns,
index=np.unique(communities_leiden)
)
leiden_clusters.index.name = 'Cluster'
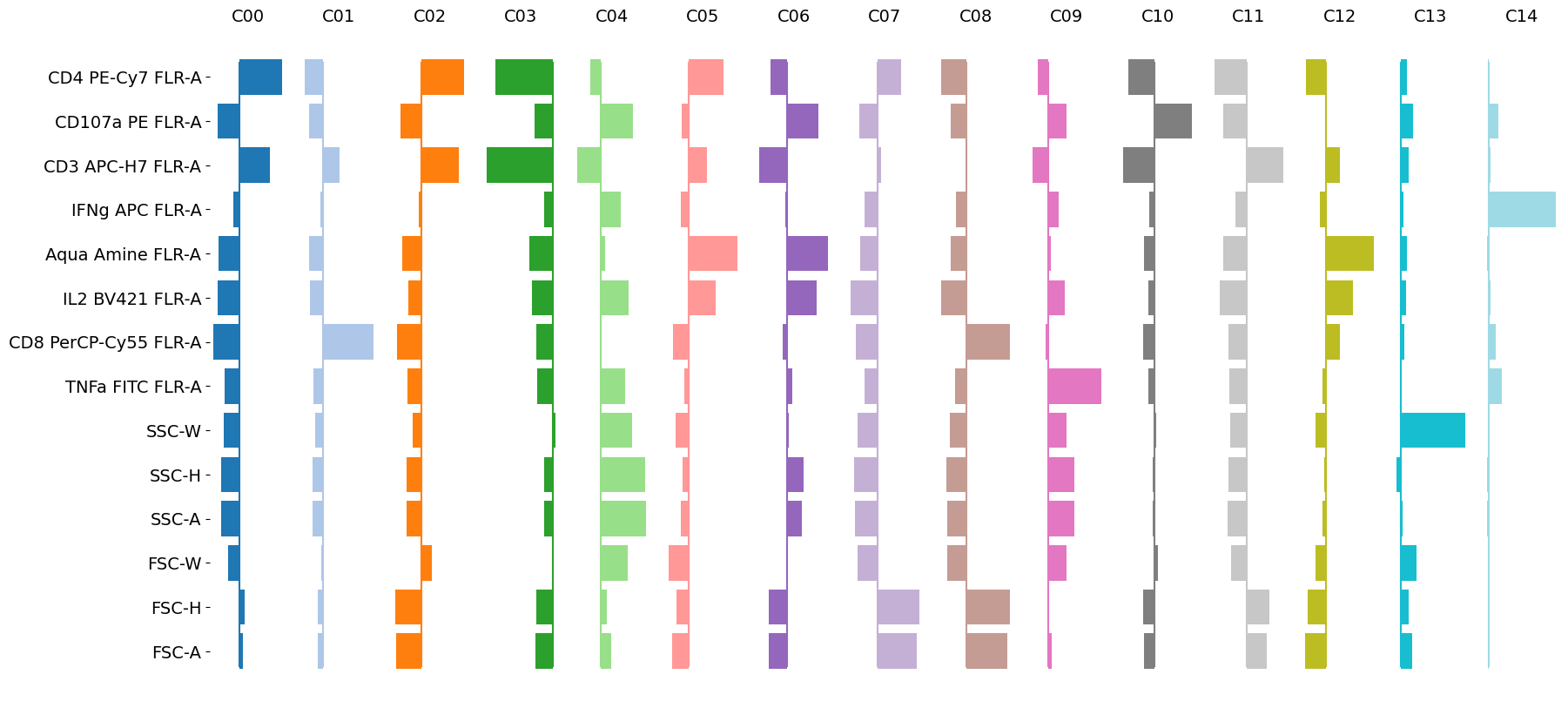
[19]:
tab20 = plt.colormaps['tab20']
n, p = leiden_clusters.shape
fig, axes = plt.subplots(1, n, figsize=(20, 10))
for i, ax in enumerate(axes.ravel()):
ax.barh(range(p), leiden_clusters.iloc[i,:], color=tab20(int(i*(20+1)/n)))
ax.set_xticks([])
ax.set_yticks([])
ax.set_frame_on(False)
ax.set_title(f'C{i:02d}', fontsize=14)
ax.axvline(0, c=tab20(int(i*(20+1)/n)), ymin=0.05, ymax=0.95)
if i == 0:
ax.set_yticks(range(p))
ax.set_yticklabels(leiden_clusters.columns,fontsize=14)
Apply dimension reduction using PaCMAP
[20]:
embedder = pacmap.PaCMAP()
[21]:
X2 = embedder.fit_transform(X_scaled)
[22]:
min_max_scaler = MinMaxScaler()
X2 = min_max_scaler.fit_transform(X2)
[23]:
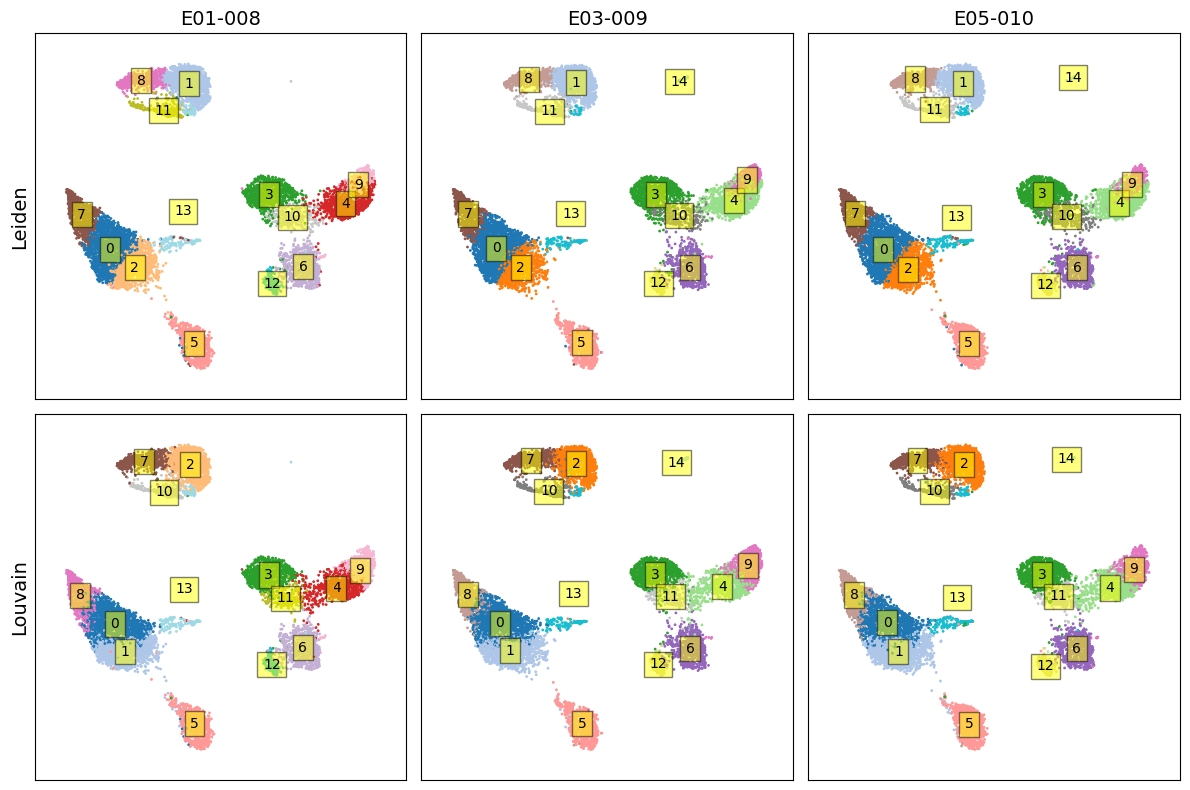
fig, axes = plt.subplots(2, 3, figsize=(12, 8))
for i, (community, title) in enumerate(zip(communities, titles)):
for j in range(3):
z = community[(j*k):(j+1)*k]
x = X2[(j*k):(j+1)*k, 0]
y = X2[(j*k):(j+1)*k, 1]
ax = axes[i, j]
ax.scatter(x, y, s=1, c=z, cmap='tab20')
ax.set_xticks([])
ax.set_yticks([])
ax.set_xlim([-0.1,1.1])
ax.set_ylim([-0.1,1.1])
if j==0:
ax.set_ylabel(title, fontsize=14)
if i==0:
ax.set_title('-'.join(sample_ids[j].split('_')[3:5]), fontsize=14)
for idx in np.unique(z):
x_, y_ = x[z==idx], y[z==idx]
x_c, y_c = np.mean(x_), np.mean(y_)
ax.text(
x_c,
y_c,
str(idx),
va='center',
ha='center',
bbox=dict(fc='yellow', alpha=0.5)
)
plt.tight_layout()
[ ]: