Saving Flow Analysis Data as AnnData objects for ScanPy
Create AnnData object from FlowJo Workspace analysis
[1]:
import os
import anndata as ad
import matplotlib.pyplot as plt
from matplotlib.pyplot import rc_context
import numpy as np
import pandas as pd
import scanpy as sc
import joypy
import flowkit as fk
[2]:
import warnings
#warnings.simplefilter('ignore', UserWarning)
# warnings.simplefilter('ignore', RuntimeWarning)
# warnings.simplefilter('ignore', FutureWarning)
# warnings.simplefilter('ignore', pd.errors.PerformanceWarning)
[3]:
# sc.settings.verbosity = 3
# sc.settings.set_figure_params(dpi=75, facecolor='white')
# sc.logging.print_header()
[4]:
base_dir = "../../../data/8_color_data_set"
sample_path = os.path.join(base_dir, "fcs_files")
wsp_path = os.path.join(base_dir, "8_color_ICS.wsp")
seed = 123
Import FlowJo workspace and process samples
[5]:
workspace = fk.Workspace(wsp_path, fcs_samples=sample_path)
[6]:
workspace.summary()
[6]:
| samples | loaded_samples | gates | max_gate_depth | |
|---|---|---|---|---|
| group_name | ||||
| All Samples | 3 | 3 | 14 | 6 |
| DEN | 3 | 3 | 14 | 6 |
| GEN | 0 | 0 | 0 | 0 |
| G69 | 0 | 0 | 0 | 0 |
| Lyo Cells | 0 | 0 | 0 | 0 |
[7]:
# The 'DEN' group has the analysis of interest
sample_group = 'DEN'
sample_ids = workspace.get_sample_ids(group_name=sample_group)
[8]:
# check the event counts for all samples (just for sanity checking our data)
for sample_id in sample_ids:
sample = workspace.get_sample(sample_id)
print(sample_id, sample.event_count)
101_DEN084Y5_15_E01_008_clean.fcs 290172
101_DEN084Y5_15_E03_009_clean.fcs 283969
101_DEN084Y5_15_E05_010_clean.fcs 285290
[9]:
workspace.analyze_samples(sample_group)
Get Boolean arrays of gate membership for all events for all gates for each sample
[10]:
# DataFrame to hold gate membership data, columns are gates, rows are events
df_gate_membership = pd.DataFrame()
# choose a sample to get the gates from, we assume all samples have the same gate tree
for gate_name, gate_path in workspace.get_gate_ids(sample_ids[0]):
results = []
for sample_id in sample_ids:
result = workspace.get_gate_membership(
sample_id,
gate_name=gate_name,
gate_path=gate_path
)
results.append(result)
results = np.concatenate(results)
df_gate_membership[':'.join(list(gate_path) + [gate_name])] = results
[11]:
df_gate_membership.head()
[11]:
| root:Time | root:Time:Singlets | root:Time:Singlets:aAmine- | root:Time:Singlets:aAmine-:CD3+ | root:Time:Singlets:aAmine-:CD3+:CD4+ | root:Time:Singlets:aAmine-:CD3+:CD4+:CD107a+ | root:Time:Singlets:aAmine-:CD3+:CD4+:IFNg+ | root:Time:Singlets:aAmine-:CD3+:CD4+:IL2+ | root:Time:Singlets:aAmine-:CD3+:CD4+:TNFa+ | root:Time:Singlets:aAmine-:CD3+:CD8+ | root:Time:Singlets:aAmine-:CD3+:CD8+:CD107a+ | root:Time:Singlets:aAmine-:CD3+:CD8+:IFNg+ | root:Time:Singlets:aAmine-:CD3+:CD8+:IL2+ | root:Time:Singlets:aAmine-:CD3+:CD8+:TNFa+ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | False | False | False | False | False | False | False | False | False | False | False | False | False | False |
| 1 | False | False | False | False | False | False | False | False | False | False | False | False | False | False |
| 2 | False | False | False | False | False | False | False | False | False | False | False | False | False | False |
| 3 | False | False | False | False | False | False | False | False | False | False | False | False | False | False |
| 4 | False | False | False | False | False | False | False | False | False | False | False | False | False | False |
[12]:
# verify our event count looks right
df_gate_membership.shape
[12]:
(859431, 14)
Get processed events for all samples
A FlowJo workspace has a set of transforms, one for each channel in a sample. However, all samples are not guaranteed to have the same set of transforms, so always good to check them.
We’ll use the get_gate_events method without specifying the gate to get processed events for the root level (i.e. all events for each sample).
[13]:
df_processed_events = []
for sample_id in sample_ids:
# The get_gate_events method returns all events if not given a gate
df = workspace.get_gate_events(sample_id)
df_processed_events.append(df)
df_processed_events = pd.concat(df_processed_events)
[14]:
df_processed_events.shape
[14]:
(859431, 16)
[15]:
df_processed_events.head()
[15]:
| sample_id | FSC-A | FSC-H | FSC-W | SSC-A | SSC-H | SSC-W | TNFa FITC FLR-A | CD8 PerCP-Cy55 FLR-A | IL2 BV421 FLR-A | Aqua Amine FLR-A | IFNg APC FLR-A | CD3 APC-H7 FLR-A | CD107a PE FLR-A | CD4 PE-Cy7 FLR-A | Time | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.669193 | 0.550243 | 0.304044 | 0.145722 | 0.136929 | 0.266054 | 0.246065 | 0.297479 | 0.280577 | 0.248555 | 0.255891 | 0.532903 | 0.300733 | 0.566413 | 0.035940 |
| 1 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.470615 | 0.405136 | 0.290405 | 0.200456 | 0.187286 | 0.267580 | 0.243213 | 0.402105 | 0.281486 | 0.245255 | 0.249686 | 0.212227 | 0.278771 | 0.244356 | 0.035983 |
| 2 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.618339 | 0.518814 | 0.297958 | 0.165320 | 0.157120 | 0.263048 | 0.236940 | 0.638401 | 0.275555 | 0.247613 | 0.247531 | 0.466408 | 0.281839 | 0.268400 | 0.036026 |
| 3 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.466790 | 0.384033 | 0.303874 | 0.136375 | 0.127686 | 0.267014 | 0.241100 | 0.646098 | 0.272339 | 0.249497 | 0.249413 | 0.433891 | 0.275977 | 0.238305 | 0.036040 |
| 4 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.321537 | 0.268814 | 0.299033 | 0.119671 | 0.111221 | 0.268994 | 0.248873 | 0.478198 | 0.328231 | 0.258179 | 0.231071 | 0.487078 | 0.328163 | 0.288373 | 0.036068 |
Create AnnData object
The initial data will be the processed events
[16]:
# event values without the sample_id column
data = df_processed_events.iloc[:, 1:].values
[17]:
adata = ad.AnnData(data)
[18]:
adata.X
[18]:
array([[0.66919339, 0.55024338, 0.30404428, ..., 0.30073305, 0.56641316,
0.03594016],
[0.47061452, 0.40513611, 0.29040521, ..., 0.27877141, 0.24435586,
0.03598285],
[0.6183387 , 0.51881409, 0.29795775, ..., 0.28183939, 0.26840002,
0.03602554],
...,
[0.49872547, 0.42063522, 0.29641208, ..., 0.27990703, 0.61764152,
0.98959426],
[0.5290575 , 0.34724426, 0.38089722, ..., 0.61813569, 0.52649112,
0.98960942],
[0.39496988, 0.33108139, 0.2982423 , ..., 0.43869763, 0.62046722,
0.98960942]])
Event labels
Basically just using an index for the event labels
[19]:
adata.obs_names = np.arange(adata.shape[0]).astype('str')
Marker labels
[20]:
adata.var_names = df_processed_events.columns[1:]
Attach sample information to each event
[21]:
adata.obs['sample_id'] = pd.Categorical(df_processed_events.sample_id)
[22]:
adata.obs.head(3)
[22]:
| sample_id | |
|---|---|
| 0 | 101_DEN084Y5_15_E01_008_clean.fcs |
| 1 | 101_DEN084Y5_15_E01_008_clean.fcs |
| 2 | 101_DEN084Y5_15_E01_008_clean.fcs |
[23]:
adata
[23]:
AnnData object with n_obs × n_vars = 859431 × 15
obs: 'sample_id'
Add Boolean matrix of gate indices
[24]:
adata.obs['sample_id'].index
[24]:
Index(['0', '1', '2', '3', '4', '5', '6', '7', '8', '9',
...
'859421', '859422', '859423', '859424', '859425', '859426', '859427',
'859428', '859429', '859430'],
dtype='object', length=859431)
[25]:
df_gate_membership.index = adata.obs['sample_id'].index
adata.obsm['gate_membership'] = df_gate_membership
[26]:
adata
[26]:
AnnData object with n_obs × n_vars = 859431 × 15
obs: 'sample_id'
obsm: 'gate_membership'
Add unstructured data for gating hierarchy and transforms
Need to convert transforms to str or cannot save to HDF5
[27]:
gate_hierarchy = {sample_id: str(workspace.get_gate_hierarchy(sample_id)) for sample_id in sample_ids}
[28]:
adata.uns['gate_hierarchy'] = gate_hierarchy
[29]:
transforms = {sample_id: str(workspace.get_transforms(sample_id)) for sample_id in sample_ids}
[30]:
adata.uns['transforms'] = transforms
Add marker summary statistics
[31]:
len(df_processed_events.columns)
[31]:
16
[32]:
df_processed_events.iloc[:10, 1:].describe()
[32]:
| FSC-A | FSC-H | FSC-W | SSC-A | SSC-H | SSC-W | TNFa FITC FLR-A | CD8 PerCP-Cy55 FLR-A | IL2 BV421 FLR-A | Aqua Amine FLR-A | IFNg APC FLR-A | CD3 APC-H7 FLR-A | CD107a PE FLR-A | CD4 PE-Cy7 FLR-A | Time | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 |
| mean | 0.513847 | 0.424834 | 0.302854 | 0.175841 | 0.162928 | 0.268044 | 0.276783 | 0.414877 | 0.299048 | 0.256286 | 0.249117 | 0.393632 | 0.335131 | 0.358282 | 0.036195 |
| std | 0.112951 | 0.095721 | 0.007951 | 0.070695 | 0.060752 | 0.006297 | 0.098275 | 0.158428 | 0.044753 | 0.017757 | 0.014083 | 0.118088 | 0.130052 | 0.147533 | 0.000199 |
| min | 0.321537 | 0.268814 | 0.290405 | 0.116014 | 0.111221 | 0.260397 | 0.236940 | 0.225618 | 0.252084 | 0.245255 | 0.231071 | 0.212227 | 0.275977 | 0.238305 | 0.035940 |
| 25% | 0.437982 | 0.350256 | 0.298274 | 0.123847 | 0.116333 | 0.264272 | 0.242302 | 0.304357 | 0.273143 | 0.248555 | 0.238181 | 0.283199 | 0.279538 | 0.259192 | 0.036029 |
| 50% | 0.492692 | 0.418480 | 0.301504 | 0.155521 | 0.147024 | 0.266997 | 0.245685 | 0.365606 | 0.281031 | 0.249967 | 0.248472 | 0.426616 | 0.292071 | 0.282596 | 0.036189 |
| 75% | 0.613940 | 0.514628 | 0.304434 | 0.200421 | 0.186127 | 0.268640 | 0.252712 | 0.544173 | 0.326186 | 0.256070 | 0.254340 | 0.481910 | 0.315959 | 0.486388 | 0.036374 |
| max | 0.669193 | 0.550243 | 0.315916 | 0.349897 | 0.309628 | 0.282515 | 0.556043 | 0.646098 | 0.397557 | 0.305441 | 0.272172 | 0.532903 | 0.701451 | 0.594672 | 0.036452 |
[33]:
df_processed_events.head(4)
[33]:
| sample_id | FSC-A | FSC-H | FSC-W | SSC-A | SSC-H | SSC-W | TNFa FITC FLR-A | CD8 PerCP-Cy55 FLR-A | IL2 BV421 FLR-A | Aqua Amine FLR-A | IFNg APC FLR-A | CD3 APC-H7 FLR-A | CD107a PE FLR-A | CD4 PE-Cy7 FLR-A | Time | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.669193 | 0.550243 | 0.304044 | 0.145722 | 0.136929 | 0.266054 | 0.246065 | 0.297479 | 0.280577 | 0.248555 | 0.255891 | 0.532903 | 0.300733 | 0.566413 | 0.035940 |
| 1 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.470615 | 0.405136 | 0.290405 | 0.200456 | 0.187286 | 0.267580 | 0.243213 | 0.402105 | 0.281486 | 0.245255 | 0.249686 | 0.212227 | 0.278771 | 0.244356 | 0.035983 |
| 2 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.618339 | 0.518814 | 0.297958 | 0.165320 | 0.157120 | 0.263048 | 0.236940 | 0.638401 | 0.275555 | 0.247613 | 0.247531 | 0.466408 | 0.281839 | 0.268400 | 0.036026 |
| 3 | 101_DEN084Y5_15_E01_008_clean.fcs | 0.466790 | 0.384033 | 0.303874 | 0.136375 | 0.127686 | 0.267014 | 0.241100 | 0.646098 | 0.272339 | 0.249497 | 0.249413 | 0.433891 | 0.275977 | 0.238305 | 0.036040 |
Optionally save to disk
At this point we could save the AnnData object to disk for later processing:
adata.write('8_color_data_set.h5ad', compression="gzip")
Apply scanpy to AnnData object
[34]:
adata.var_names
[34]:
Index(['FSC-A', 'FSC-H', 'FSC-W', 'SSC-A', 'SSC-H', 'SSC-W', 'TNFa FITC FLR-A',
'CD8 PerCP-Cy55 FLR-A', 'IL2 BV421 FLR-A', 'Aqua Amine FLR-A',
'IFNg APC FLR-A', 'CD3 APC-H7 FLR-A', 'CD107a PE FLR-A',
'CD4 PE-Cy7 FLR-A', 'Time'],
dtype='object')
[35]:
# Only keep fluroescence intensity markers
exclude = ['FSC-A', 'FSC-H', 'FSC-W', 'SSC-A', 'SSC-H', 'SSC-W', 'Time']
adata = adata[:, ~adata.var_names.isin(exclude)]
[36]:
markers = [
'Aqua Amine FLR-A',
'CD3 APC-H7 FLR-A',
'CD4 PE-Cy7 FLR-A',
'CD8 PerCP-Cy55 FLR-A',
'TNFa FITC FLR-A',
'IL2 BV421 FLR-A',
'IFNg APC FLR-A',
'CD107a PE FLR-A',
]
[37]:
_ = joypy.joyplot(
adata.to_df().sample(10_000),
figsize=(8,4),
column = markers,
x_range=[0, 1],
colormap=plt.cm.autumn_r,
overlap=1.5
)
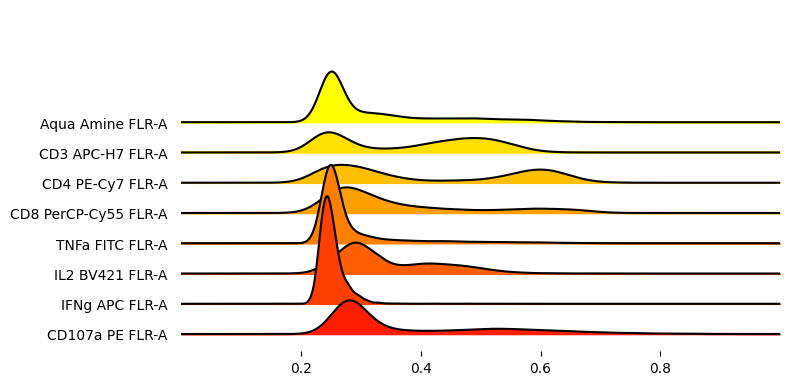
[38]:
# just to reference indices of gate names
pd.Series(adata.obsm['gate_membership'].columns)
[38]:
0 root:Time
1 root:Time:Singlets
2 root:Time:Singlets:aAmine-
3 root:Time:Singlets:aAmine-:CD3+
4 root:Time:Singlets:aAmine-:CD3+:CD4+
5 root:Time:Singlets:aAmine-:CD3+:CD4+:CD107a+
6 root:Time:Singlets:aAmine-:CD3+:CD4+:IFNg+
7 root:Time:Singlets:aAmine-:CD3+:CD4+:IL2+
8 root:Time:Singlets:aAmine-:CD3+:CD4+:TNFa+
9 root:Time:Singlets:aAmine-:CD3+:CD8+
10 root:Time:Singlets:aAmine-:CD3+:CD8+:CD107a+
11 root:Time:Singlets:aAmine-:CD3+:CD8+:IFNg+
12 root:Time:Singlets:aAmine-:CD3+:CD8+:IL2+
13 root:Time:Singlets:aAmine-:CD3+:CD8+:TNFa+
dtype: object
[39]:
cd4_funcs = adata.obsm['gate_membership'].iloc[:, [5,6,7,8]]
cd4_bool = cd4_funcs.apply(lambda xs: np.sum([x*2**i for (i, x) in enumerate(xs)]), axis=1)
cd4_num_funcs = cd4_funcs.apply(lambda xs: xs.sum(), axis=1)
[40]:
adata.obs['cd4'] = pd.Categorical(adata.obsm['gate_membership'].iloc[:, 4].astype(int).astype('str'))
adata.obs['cd4_bool'] = pd.Categorical(cd4_bool.astype('str').str.zfill(2))
adata.obs['cd4_funcs'] = pd.Categorical(cd4_num_funcs.astype('str'))
/tmp/ipykernel_1951574/3070510497.py:1: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
adata.obs['cd4'] = pd.Categorical(adata.obsm['gate_membership'].iloc[:, 4].astype(int).astype('str'))
[41]:
cd8_funcs = adata.obsm['gate_membership'].iloc[:, [10,11,12,13]]
cd8_bool = cd8_funcs.apply(lambda xs: np.sum([x*2**i for (i, x) in enumerate(xs)]), axis=1)
cd8_num_funcs = cd8_funcs.apply(lambda xs: xs.sum(), axis=1)
[42]:
adata.obs['cd8'] = pd.Categorical(adata.obsm['gate_membership'].iloc[:, 9].astype('int').astype('str'))
adata.obs['cd8_bool'] = pd.Categorical(cd8_bool.astype('str').str.zfill(2))
adata.obs['cd8_funcs'] = pd.Categorical(cd8_num_funcs.astype('str'))
Computing the neighborhood graph
To speed up UMAP calculations,
pip install pynndescent
[43]:
# Subsample to speed things up
data = sc.pp.subsample(adata, n_obs=50_000, copy=True)
[44]:
%%time
sc.pp.neighbors(data, n_neighbors=10, n_pcs=0)
CPU times: user 15.6 s, sys: 54.4 ms, total: 15.7 s
Wall time: 15.4 s
[45]:
%%time
sc.tl.leiden(data, resolution=0.1, flavor='leidenalg')
<timed eval>:1: FutureWarning: In the future, the default backend for leiden will be igraph instead of leidenalg.
To achieve the future defaults please pass: flavor="igraph" and n_iterations=2. directed must also be False to work with igraph's implementation.
CPU times: user 7 s, sys: 51.2 ms, total: 7.05 s
Wall time: 7.03 s
[46]:
%%time
sc.tl.paga(data)
CPU times: user 358 ms, sys: 12.7 ms, total: 371 ms
Wall time: 370 ms
[47]:
sc.pl.paga(data)
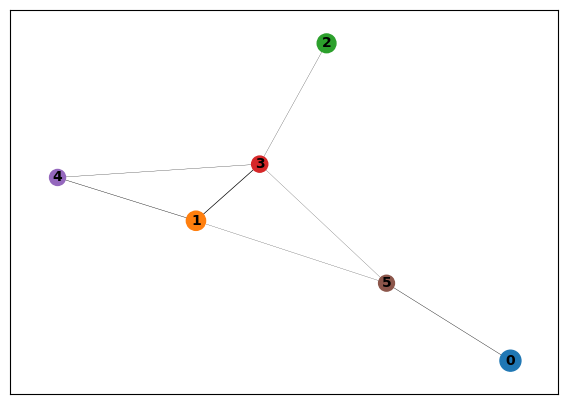
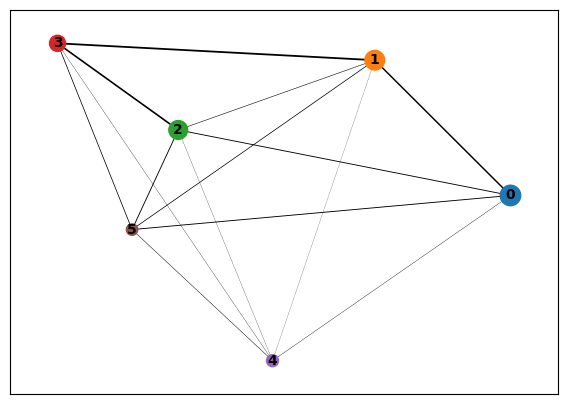
By quantifying the connectivity of partitions (groups, clusters) of the single-cell graph, partition-based graph abstraction (PAGA) generates a much simpler abstracted graph (PAGA graph) of partitions, in which edge weights represent confidence in the presence of connections. By thresholding this confidence in :func:~scanpy.pl.paga, a much simpler representation of the manifold data is obtained, which is nonetheless faithful to the topology of the manifold.
[48]:
%%time
sc.tl.umap(data, init_pos='paga')
CPU times: user 11 s, sys: 416 µs, total: 11 s
Wall time: 11 s
[49]:
%%time
sc.pl.umap(data, color=['leiden'] + markers)
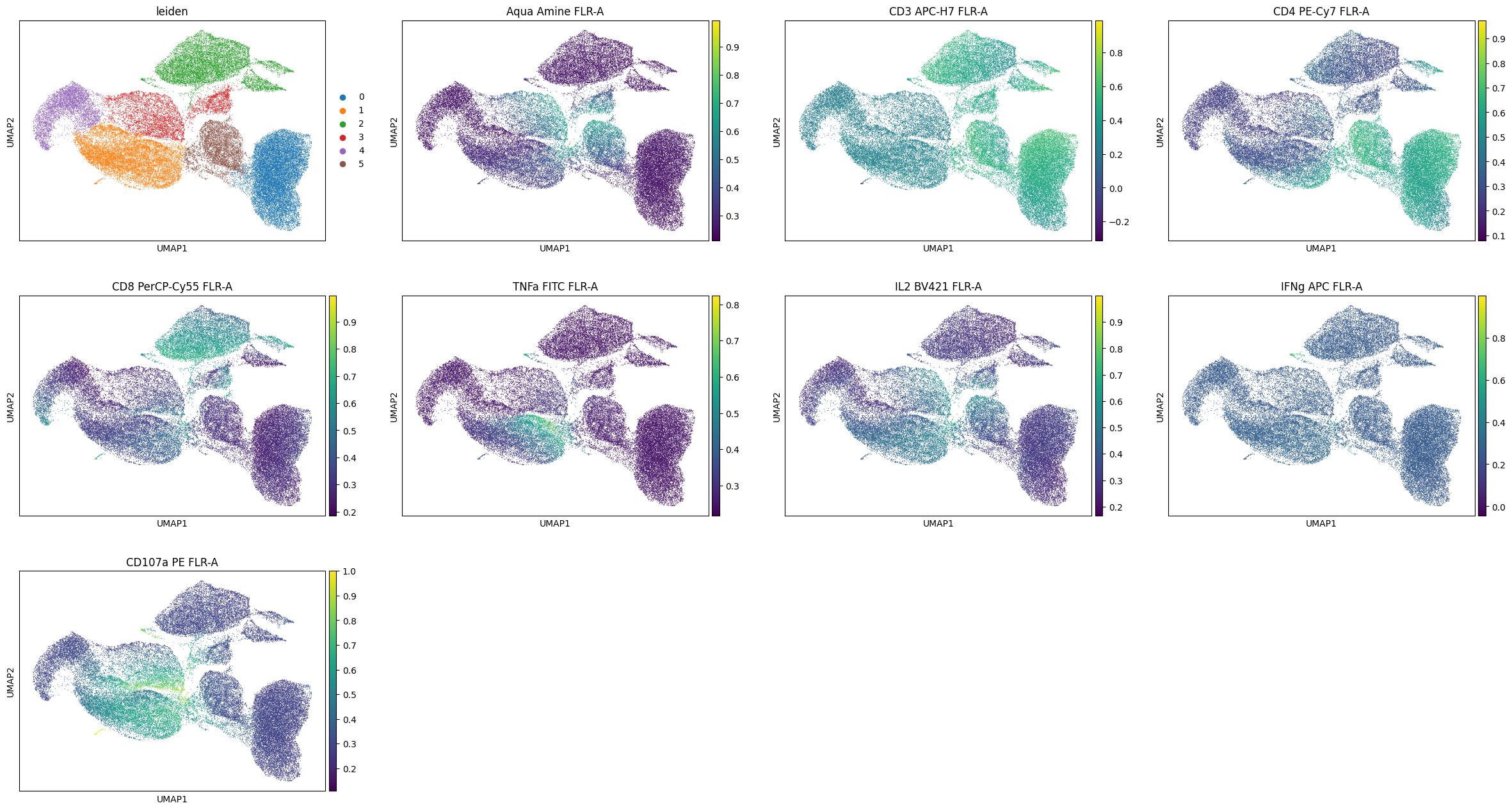
CPU times: user 1.13 s, sys: 136 ms, total: 1.27 s
Wall time: 1.09 s
[50]:
%%time
sc.pl.umap(data, color=['cd4', 'cd4_funcs', 'cd4_bool'], cmap='turbo')
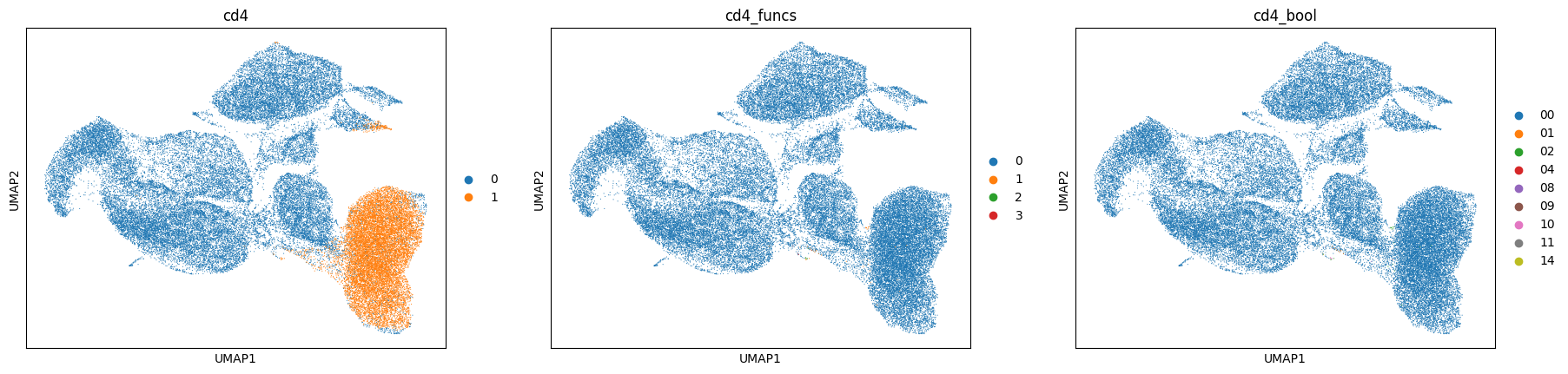
CPU times: user 454 ms, sys: 152 ms, total: 607 ms
Wall time: 422 ms
[51]:
sc.pl.umap(data, color=['cd8', 'cd8_funcs', 'cd8_bool'], cmap='turbo')
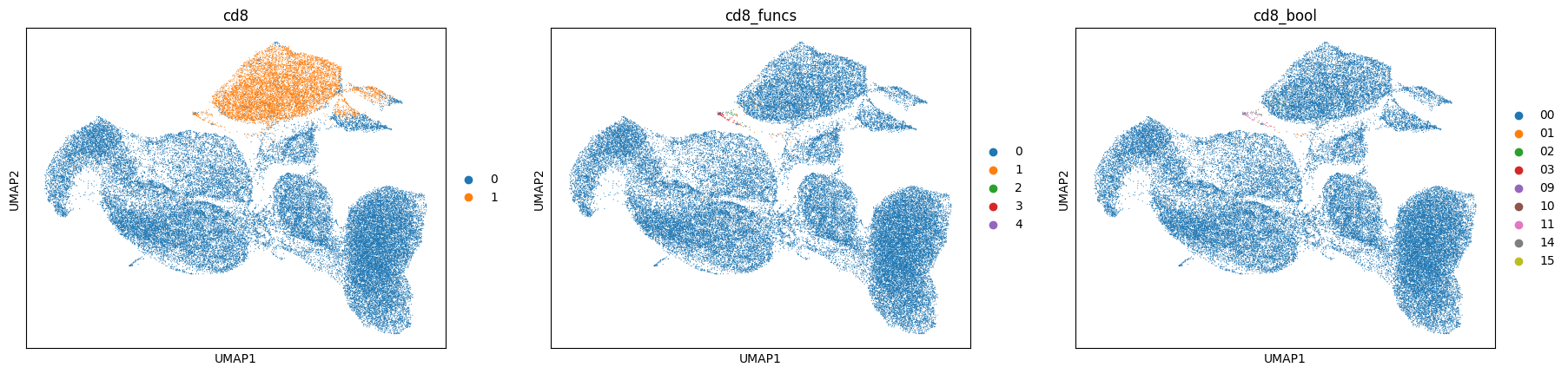
[52]:
sc.tl.embedding_density(
data,
groupby='cd4'
)
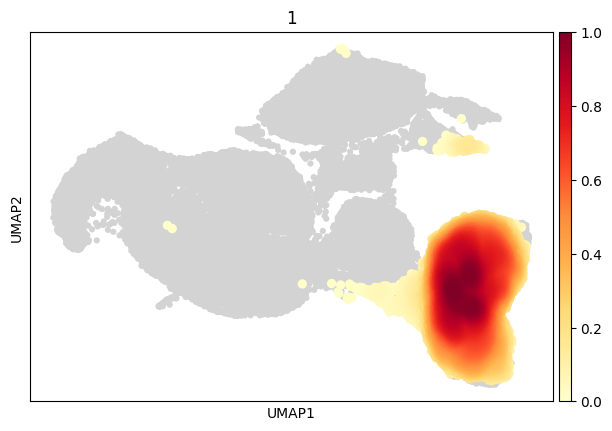
[53]:
sc.pl.embedding_density(
data,
groupby='cd4',
group='1'
)
[54]:
sc.tl.embedding_density(
data,
groupby='cd4_funcs'
)
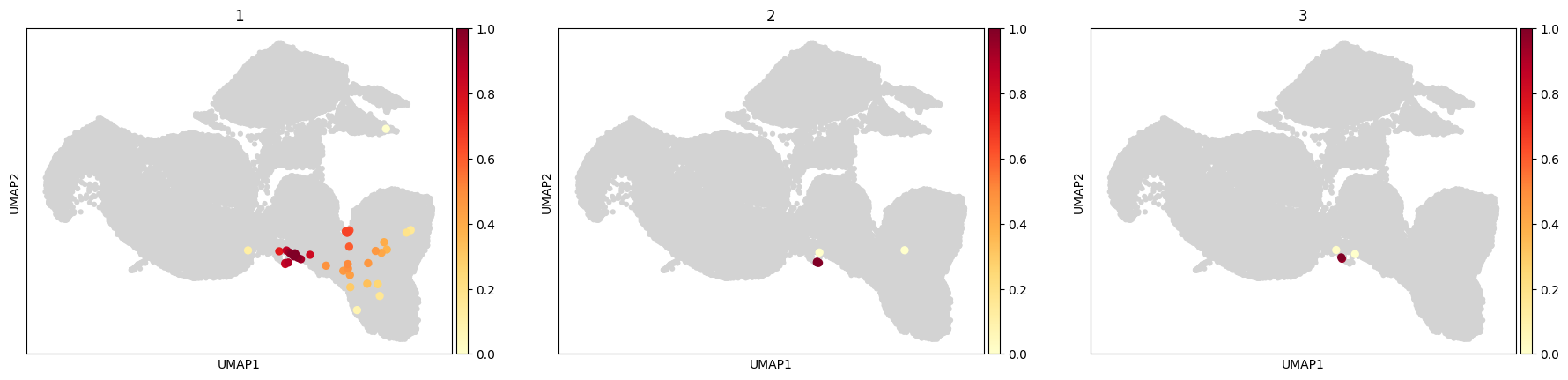
[55]:
sc.pl.embedding_density(
data,
groupby='cd4_funcs',
group=['1','2','3']
)
[56]:
sc.tl.embedding_density(
data,
groupby='cd8'
)
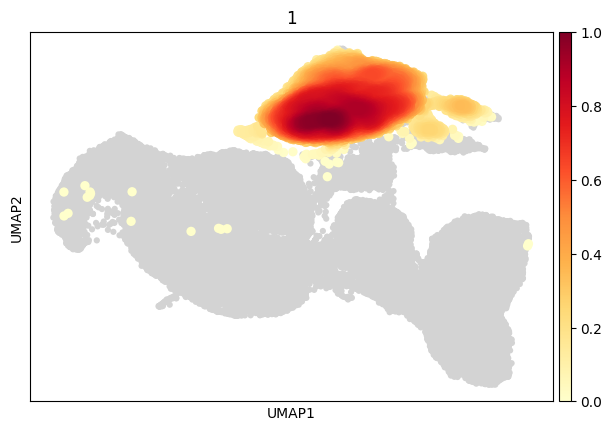
[57]:
sc.pl.embedding_density(
data,
groupby='cd8',
group='1'
)
[58]:
sc.tl.embedding_density(
data,
groupby='cd8_funcs',
)
[59]:
sc.pl.embedding_density(
data,
groupby='cd8_funcs',
group=['1', '2', '3', '4'],
)
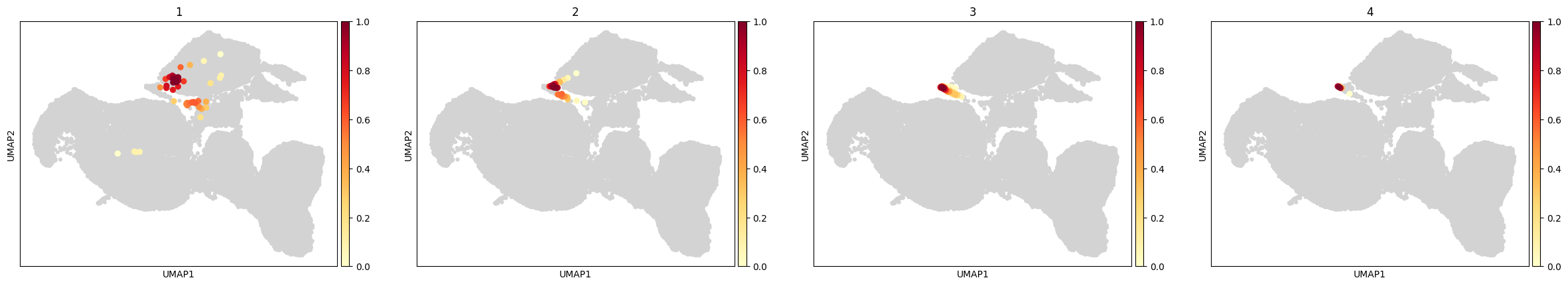
Next example: Repeat but for CD8 T cells only
[60]:
cd8_data = adata[adata.obsm['gate_membership']['root:Time:Singlets:aAmine-:CD3+:CD8+']]
[61]:
cd8_data
[61]:
View of AnnData object with n_obs × n_vars = 140676 × 8
obs: 'sample_id', 'cd4', 'cd4_bool', 'cd4_funcs', 'cd8', 'cd8_bool', 'cd8_funcs'
uns: 'gate_hierarchy', 'transforms'
obsm: 'gate_membership'
[62]:
sc.pp.neighbors(cd8_data, n_neighbors=30, n_pcs=0)
[63]:
sc.tl.leiden(cd8_data, resolution=0.2, flavor='leidenalg')
[64]:
sc.tl.paga(cd8_data)
[65]:
sc.pl.paga(cd8_data)
[66]:
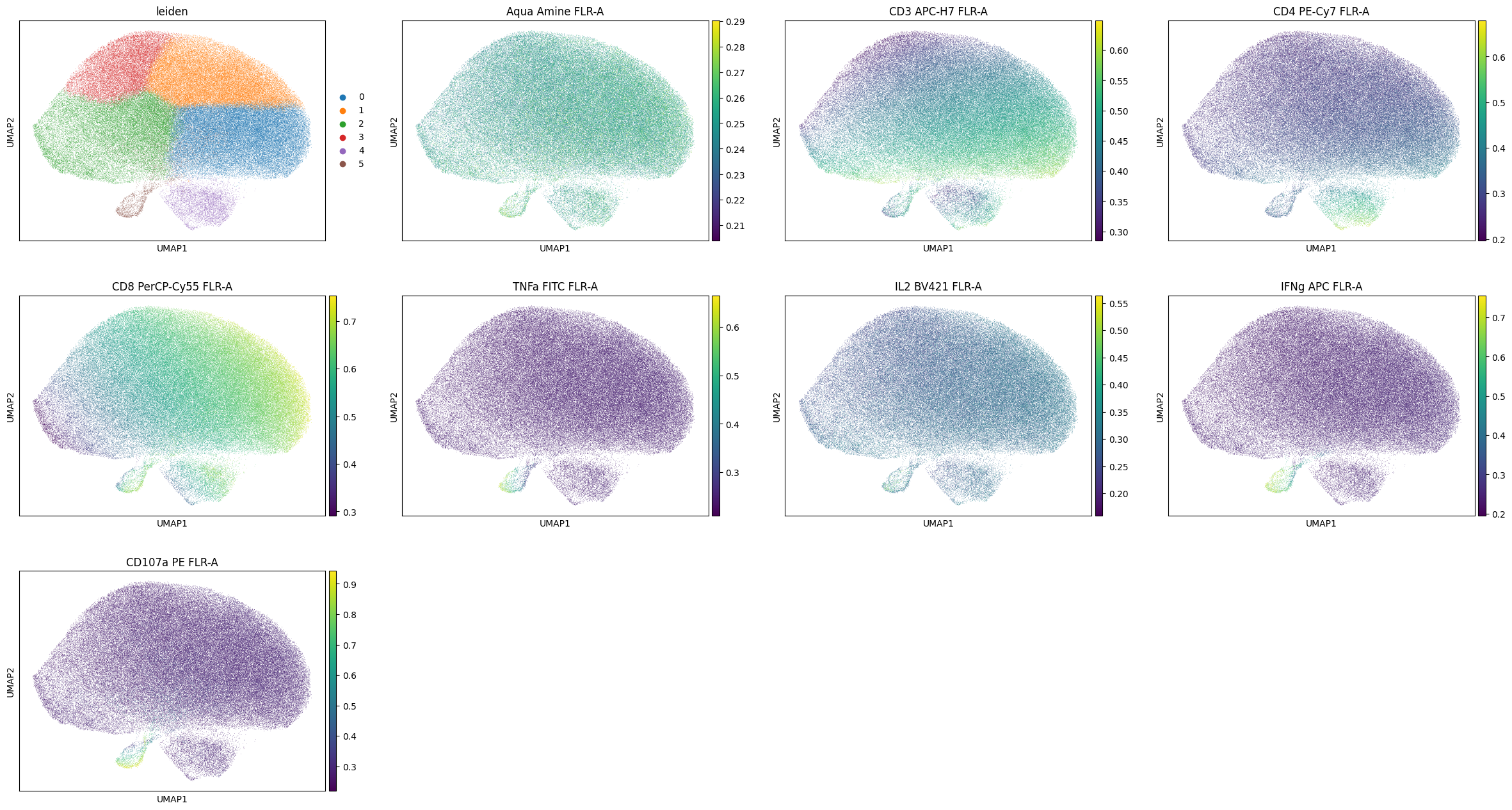
sc.tl.umap(cd8_data, init_pos='paga')
[67]:
sc.pl.umap(
cd8_data,
color=['leiden'] + markers,
)
[68]:
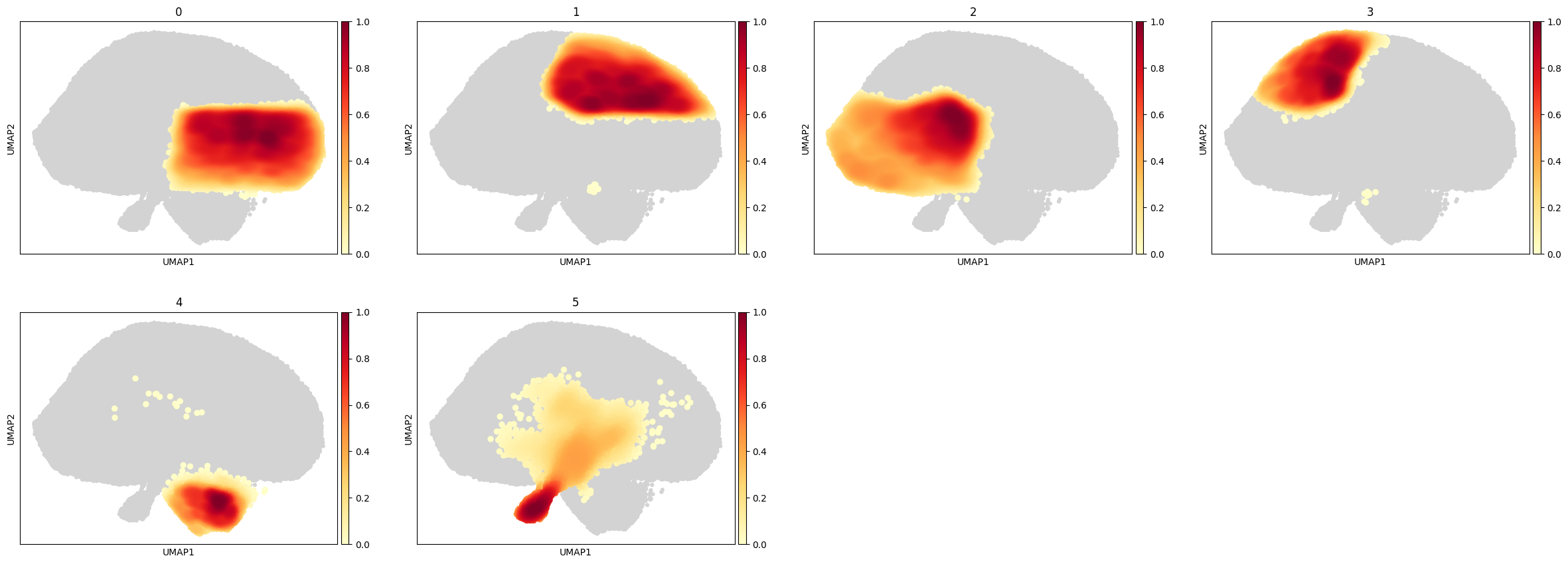
sc.tl.embedding_density(
cd8_data,
groupby='leiden'
)
[69]:
sc.pl.embedding_density(
cd8_data,
groupby='leiden',
)
Finding markers
[70]:
sc.tl.rank_genes_groups(
cd8_data,
groupby='leiden',
method='wilcoxon'
)
[71]:
sc.pl.violin(cd8_data, markers, groupby='leiden')

[72]:
sc.pl.dotplot(cd8_data, markers, groupby='leiden', swap_axes=True)
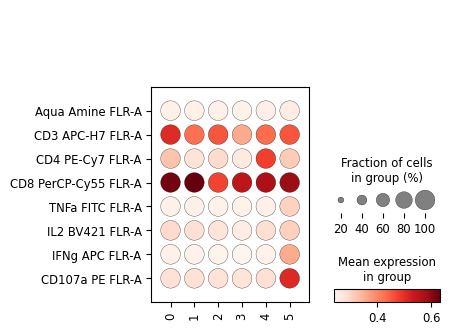
[73]:
sc.pl.stacked_violin(cd8_data, markers, groupby='leiden', swap_axes=True)
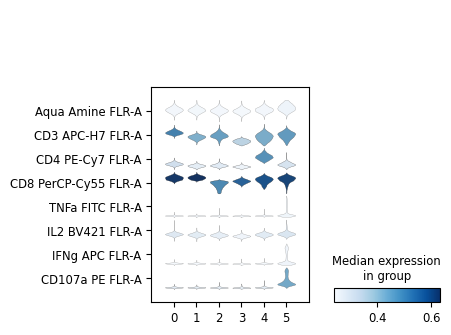
[74]:
sc.tl.dendrogram(cd8_data, groupby='leiden')
[75]:
sc.pl.matrixplot(
cd8_data,
markers,
groupby='leiden',
swap_axes=True,
standard_scale='var',
log=False,
dendrogram=True,
)
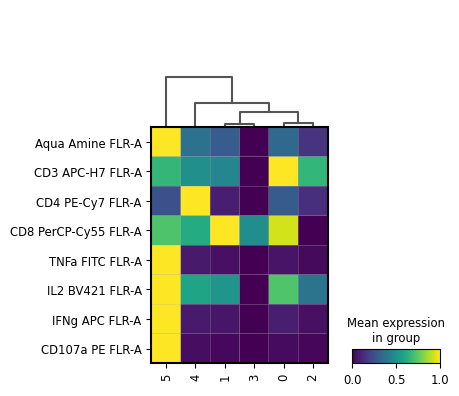
[76]:
sc.pl.heatmap(
cd8_data,
markers,
groupby='leiden',
swap_axes=True,
figsize=(12,4)
)
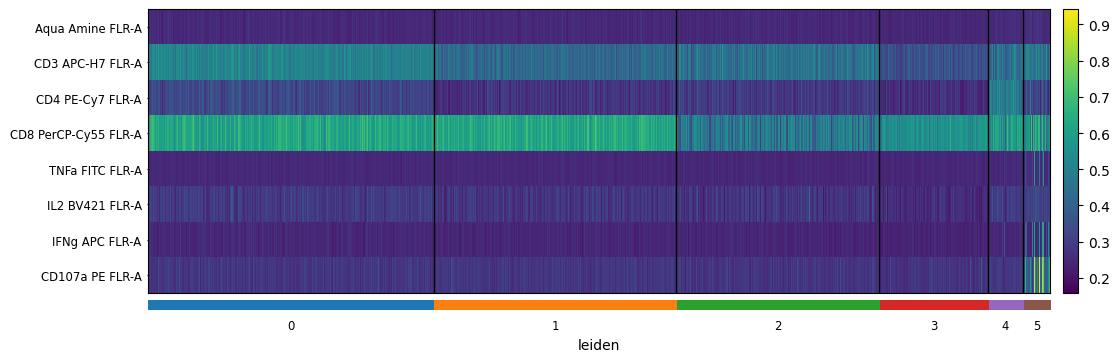
[77]:
sc.pl.tracksplot(
cd8_data,
markers,
groupby='leiden',
swap_axes=True,
figsize=(12,4)
)
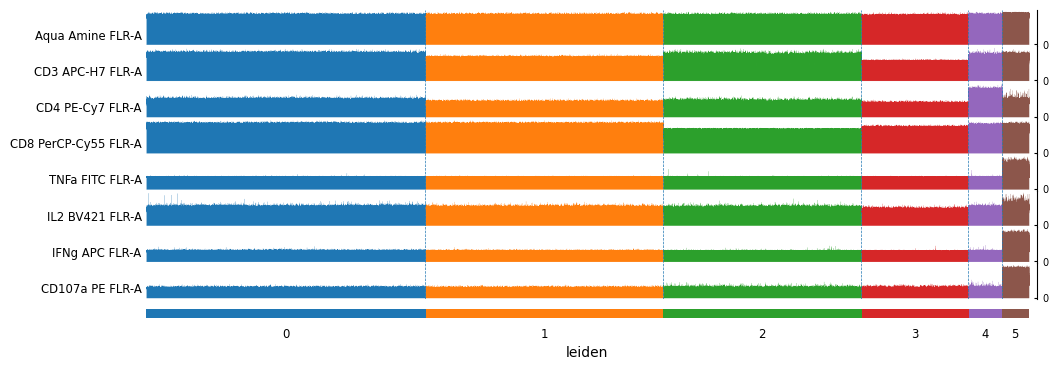
[78]:
cd8_data.layers['scaled'] = sc.pp.scale(cd8_data, copy=True).X
[79]:
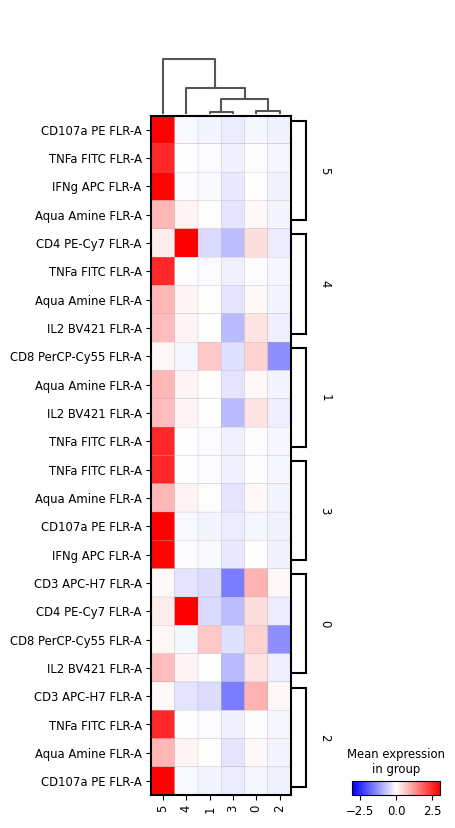
sc.pl.rank_genes_groups_matrixplot(
cd8_data,
n_genes=4,
use_raw=False,
vmin=-3,
vmax=3,
cmap='bwr',
layer='scaled',
swap_axes=True
)
[80]:
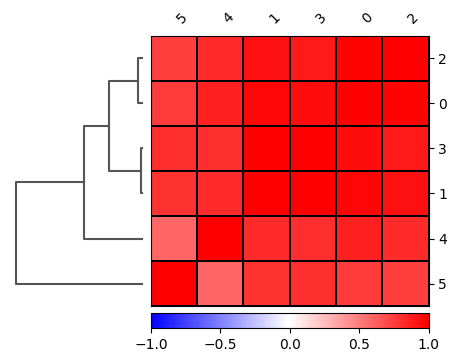
sc.pl.correlation_matrix(
cd8_data,
groupby='leiden',
)
[ ]: